def expand_neighbors_impl(ghCenter: GeoHash, ghCur: GeoHash, buffer: collection.mutable.Set[GeoHash]): Unit = {
// MARK: DP: check whether it's iterated already or not
if(buffer contains ghCur) {
return
}
buffer += ghCur
for(ghAround <- get4GeoHashAround(ghCur)) {
if(distanceBetweenGeohash(ghCenter, ghAround) <= radius) {
expand_neighbors_impl(ghCenter, ghAround, buffer)
}
}
}
def get4GeoHashAround(gh: GeoHash): Array[GeoHash] = {
Array(gh.getNorthernNeighbour, gh.getSouthernNeighbour, gh.getWesternNeighbour, gh.getEasternNeighbour)
}
def distanceBetweenGeohash(gh1: GeoHash, gh2: GeoHash) = {
haversine(gh1.getBoundingBoxCenterPoint.getLatitude, gh1.getBoundingBoxCenterPoint.getLongitude, gh2.getBoundingBoxCenterPoint.getLatitude, gh2.getBoundingBoxCenterPoint.getLongitude)
}
@tailrec
def expand_neighbors_impl(ghCenter: GeoHash, toGoThrough: List[GeoHash], buffer: Set[GeoHash] = Set()): Set[GeoHash] = {
toGoThrough.headOption match {
case None => buffer
case Some(ghCur) =>
if (buffer contains ghCur) {
expand_neighbors_impl(ghCenter, toGoThrough.tail, buffer)
}
else {
val neighbors = get4GeoHashAround(ghCur).filter(distanceBetweenGeohash(ghCenter, _) <= radius)
expand_neighbors_impl(ghCenter, neighbors ++: toGoThrough, buffer + ghCur)
}
}
}
def expand_neighbors(ghCenter: GeoHash, ghCur: GeoHash): Set[GeoHash] = expand_neighbors_impl(ghCenter, List(ghCur))
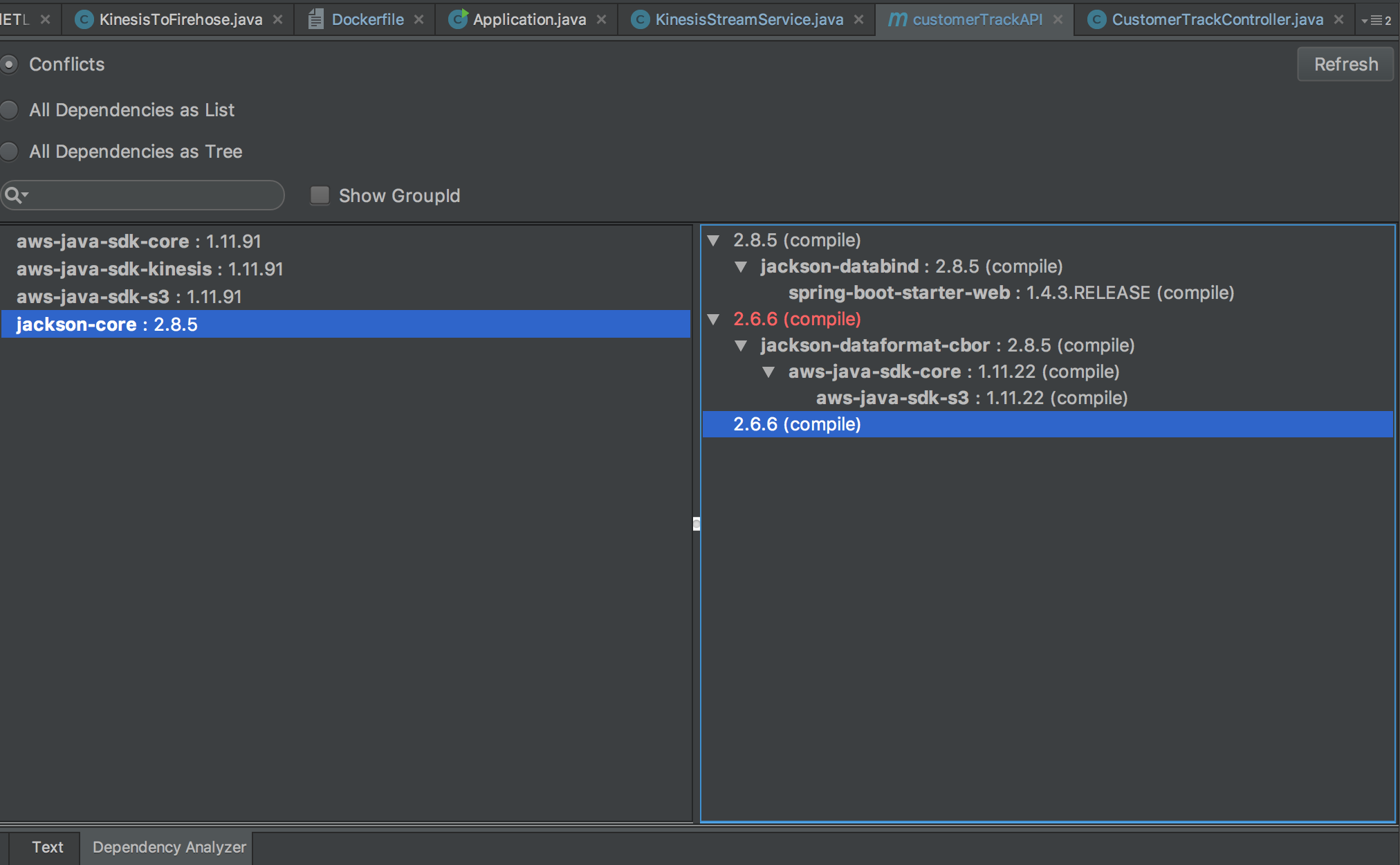
GeoHash. The following will start searching from a grid, whose path will be one of the (north, south, east, west) grids. The border will be distance between current grid and the starting grid.